Epidemic Modeling
Especially at the start of an outbreak, mitigating the spread of infectious diseases is an incredibly time sensitive task, as cases tend to rise exponentially during the initial stages. Despite this, it is often hard to immediately evaluate the efficacy of any given intervention, as the effects of changes in policy are not felt until weeks after they are implemented. One major reason for this is the lag time between when someone is infected and when they are potentially diagnosed, hospitalized, or killed. Together, these two issues mean that responding to an outbreak requires public health officials to make quick decisions based on limited information.
Agent-Based Modeling
One way of mitigating this issue is to use computational models to predict the effects of any proposed interventions. Traditional compartmental models for the spread of infectious diseases rely on coupled rate equations - systems of differential equations relating the number of people who are susceptible, exposed, infected, and recovered (SEIR). However, while such models are useful for capturing population-level trends, they fail to capture the low-level behavior that many interventions target.
Another class of models, termed agent-based models, are well-suited for modeling this sort of individual behavior. In an agent-based model, individual members of the population are simulated directly, not in aggregate. This makes modeling the impact of interventions that only indirectly impact infections - such as stay at home orders or school closures - much more straightforward, as the individual behavioral changes caused by the intervention are directly captured by the model.
Loimos
We put these agent-based modeling techniques to use in Loimos. Loimos is a highly scalable parallel code written in Charm++ which uses agent-based modeling to simulate disease spread over large, realistic, networks of interaction. These networks are given in the form of activity schedules for each simulated individual; on a given simulation day, each person will visit several locations and potentially come into contact with other people who are there at the same time (as determined by a discrete event simulation at the location). These interactions have the potential to result in infections if one person is infectious and the other is susceptible to infection. Once a person is infected, they transition through different stages of the disease over time - with changing levels of infectivity, symptoms, and so forth - until they either recover or die. By tracking how many people in in each disease state over the course of the simulation, we can calculate cases, hospitalizations, and deaths over the course of the simulated outbreak at the county, state, and national levels.
We have been able to demonstrate that our implementation of Loimos is able to scale to large core counts on different HPC platforms such as Perlmutter and Theta, at NERSC and ALCF respectively. In particular, Loimos is able to simulate the spread of COVID-19 on a variety of realisttic digital twin populations representing US states. These datasets have been developed by our collaborators at the Biocomplexity Institude at the University of Virginia.
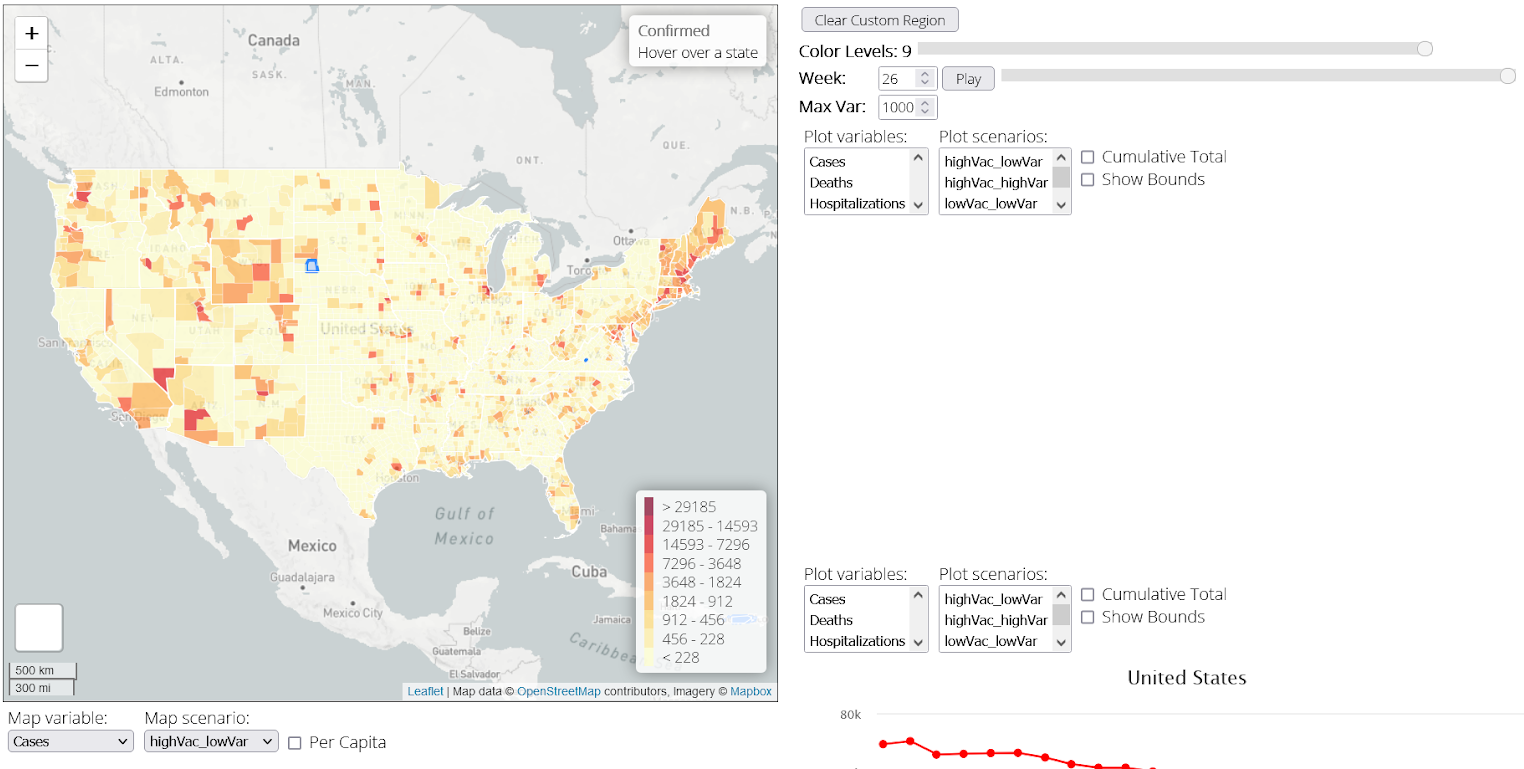
We have also been developing a tool to help visualize the outcomes of
our simulations. Currently, this tool can display county, state, and
national case, hospitalization, and death data for a given simulation
run. The visualization may be seen below.